
40
Department of Human Genetics, McGill University, Montreal, QC H3Z 2Z3, Canada
41
These authors contributed equally to this work
42
These authors contributed equally to this work
*Correspondence: c.plass@dkfz.de (C.P.), nada.jabado@mcgill.ca (N.J.), s.pfister@dkfz.de (S.M.P.)
http://dx.doi.org/10.1016/j.ccr.2012.08.024
SUMMARY
Glioblastoma (GBM) is a brain tumor that carries a dismal prognosis and displays considerable heteroge-
neity. We have recently identified recurrent H3F3A mutations affecting two critical amino acids (K27 and
G34) of histone H3.3 in one-third of pediatric GBM. Here, we show that each H3F3A mutation defines an
epigenetic subgrou p of GBM with a distinct global methylation pattern, and that they are mutually exclusive
with IDH1 mutations, which characteriz e a third mutation-defined subgroup. Three further epigenetic
subgroups were enr iched for hallmark genetic events of adult GBM and/or established transcriptomic signa-
tures. We also demonstrate that the two H3F3A mutations give rise to GBMs in separate anatomic compart-
ments, with differential regulation of transcription factors OLIG1, OLIG2, and FOXG1, possibly reflecting
different cellular origins.
INTRODUCTION
Glioblastoma (GBM; World Health Organization [WHO] grade IV),
the most common primary brain tumor, carries a universally
dismal prognosis in children and adults (Louis et al., 2007).
With evidence emerging recently of age-specific molecular and
genetic differences, it is now becoming apparent that pediatric
GBM is largely biologically distinct from adult GBM. Based on
similarities in recurrent genomic aberrations (Bax et al., 2010;
McLendon et al., 2008; Paugh et al., 2010; Qu et al., 2010; Schiff-
man et al., 2010; Zarghooni et al., 2010), it was long thought
that pediatric GBMs more closely resembled adult ‘‘secondary’’
GBM, which arise from a preceding lower-grade lesion. How-
ever, stepwise transformation from less-malignant gliomas into
GBM rarely occurs in children (Broniscer et al., 2007). Further-
more, IDH1 or IDH2 mutations, which are found in up to 98%
of adult secondary GBM, are very rare in childhood GBM
(<10%) (Antonelli et al., 2010; Balss et al., 2008; De Carli et al.,
2009; Paugh et al., 2010; Pollack et al., 2011; Schiffman et al.,
2010; Setty et al., 2010; Yan et al., 2009).
We recently identified two recurrent somatic mutations in the
H3F3A gene, affecting highly conserved residues of its encoded
protein, the replication-independent histone 3 variant H3.3, in
one-third of pediatric GBMs ( Schwartzentruber et al., 2012).
Mutations in a protein complex comprised of H3.3 and ATRX/
DAXX were detected in 45% of cases, and were shown to be
associated with TP53 mutations and alternative lengthening of
telomeres (ALT). The H3.3 mutations result in amino acid sub-
stitutions at K27 or G34—at or near residues targeted by key
post-translational modifications that regulate H3.3’s activity in
governing gene expression (Hyland et al., 2011)—and were
shown to be linked to distinct transcriptional profiles (Schwart-
zentruber et al., 2012). Methylation of K27 and K36 is also disrup-
ted by elevated levels of the onco-metabolite 2-hydroxyglutarate
(2-HG) resulting from gain-of-function mutations in IDH1
(Chowdhury et al., 2011; Xu et al., 2011), which was previously
shown to be associated with a distinct Glioma-CpG-Island
Methylator Phenotype (G-CIMP) (Noushmehr et al., 2010).
In the present study, we further investigate the heterogeneity
of glioblastoma across the entire age spectrum, and elucidate
the impact of H3F3A mutations on the GBM epigenome.
RESULTS
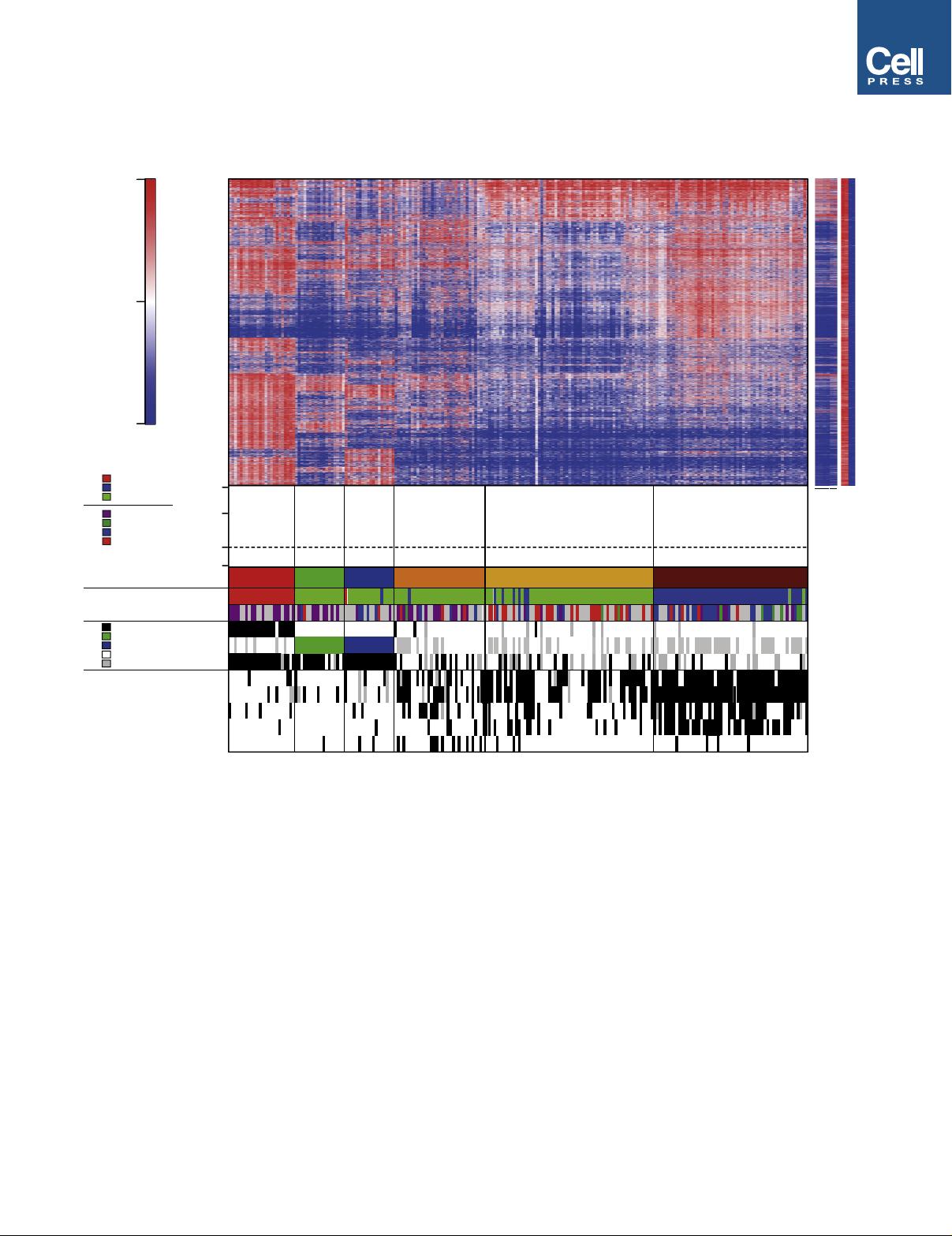
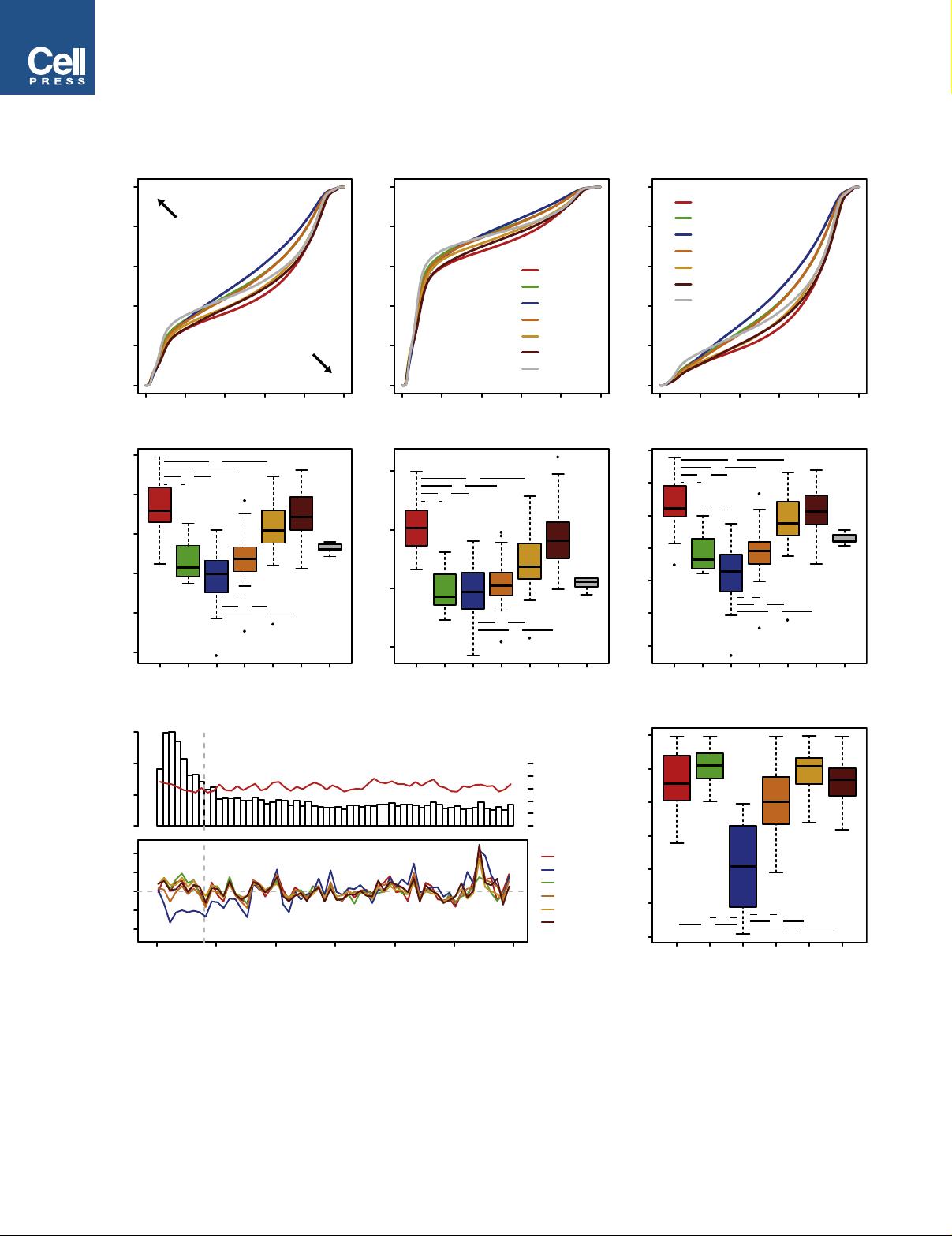
Integrated Molecular Classification of Glioblastoma
We used an integrative approach based on epigenetic, copy-
number, expression, and genetic analyses to investigate the
heterogeneity of glioblastoma across all age groups. An over-
view of all GBM samples subjected to various analyses is given
in Figure S1A available online.
Significance
GBM is the most common and also the most devastating brain tumor, with a 5-year survival rate below 10%. We present
strong evidence that GBM can be subclassified into multiple groups, indistinguishable by histological appearance, but
correlating with molecular-genetic factors as well as key clinical variables such as patient age and tumor location. We iden-
tified six epigenetic GBM subgroups displaying characteristic global DNA methylation patterns, harboring distinct hotspot
mutations, DNA copy-number alterations, and transcriptomic patterns. These findings may guide the identification of inno-
vative subgroup-specific treatments, e.g., targeted epigenetic therapies for H3.3-mutated variants, and improve the design
of future clinical trials. Our study enables classification of GBM across the entire age continuum into biologically meaningful
subgroups carrying clinical implications.
Cancer Cell
Epigenetic and Biological Subgroups of Glioblastoma
426 Cancer Cell 22, 425–437, October 16, 2012 ª2012 Elsevier Inc.


 03_complexheatmap.7z (6个子文件)
03_complexheatmap.7z (6个子文件)  data
data  450K_annotation.txt 17KB
450K_annotation.txt 17KB pc_tx_tss.bed 2.17MB
pc_tx_tss.bed 2.17MB .Rhistory 17KB
.Rhistory 17KB correction.Rproj 218B
correction.Rproj 218B PIIS1535610812003649.pdf 2.2MB
PIIS1535610812003649.pdf 2.2MB 01_complexheatmap.R 6KB
01_complexheatmap.R 6KB
 我的内容管理
展开
我的内容管理
展开
 我的资源
快来上传第一个资源
我的资源
快来上传第一个资源
 我的收益 登录查看自己的收益
我的收益 登录查看自己的收益 我的积分
登录查看自己的积分
我的积分
登录查看自己的积分
 我的C币
登录后查看C币余额
我的C币
登录后查看C币余额
 我的收藏
我的收藏  我的下载
我的下载  下载帮助
下载帮助 
 前往需求广场,查看用户热搜
前往需求广场,查看用户热搜

 信息提交成功
信息提交成功
评论0