没有合适的资源?快使用搜索试试~ 我知道了~
配体结合和核受体进化 配体结合和核受体进化 Hector Escriva、Franck Delaunay 和 Vincent Laudet* 总结 核受体形成配体激活的转录因子超家族,调节后生动物的各种生理功能,从发育到稳态。 该超家族不仅包含已知配体的受体,还包含大量所谓的孤儿受体,其配体不存在或尚未鉴定。 核受体配体结合能力的进化可能涉及孤儿受体的二次丧失,或配体受体配体结合能力的进化获得。 在这篇综述中,我们提出了来自系统发育、功能和结构研究的论点,这些研究支持在后生动物进化过程中核受体的配体结合能力有几个独立增益的假设。 BioEssays 22:717±727, 2000. ß 2000 John Wiley & Sons, Inc. 介绍 核激素受体 (NRs) 是后生动物中最丰富的一类转录调节因子,它们调节多种功能,如繁殖、分化、发育、代谢、变态和体内平衡。 NRs 作为配体激活的转录因子发挥作用,从而在控制这些过程的信号分子与转录React之间提供直接联系。 (1) NR 信号通路的基本概念涉及信号(内分泌、旁分泌、自分泌甚至内分泌)的产生、其向外周器官的转运、
资源推荐
资源详情
资源评论

Ligand binding and nuclear
receptor evolution
Hector Escriva, Franck Delaunay, and Vincent Laudet*
Summary
Nuclear receptors form a superfamily of ligand-activated
transcription factors that regulate various physiological
functions, from development to homeostasis, in me-
tazoans. The superfamily contains not only receptors
for known ligands but also a large number of so-called
orphan receptors for which ligands do not exist or have
not been identified. The evolution of ligand-binding
capacity of nuclear receptors may involve either sec-
ondary loss in orphan receptors, or evolutionary acqu-
isition of ligand-binding capacity in liganded receptors.
In this review, we present arguments from phylogenetic,
functional and structural studies that support the
hypothesis that there have been several independent
gains of ligand-binding ability of nuclear receptors
during metazoan evolution. BioEssays 22:717±727,
2000. ß 2000 John Wiley & Sons, Inc.
Introduction
Nuclear hormone receptors (NRs) are one of the most
abundant classes of transcriptional regulators in metazoans,
in which they regulate functions as diverse as reproduction,
differentiation, development, metabolism, metamorphosis
and homeostasis. NRs function as ligand-activated transcrip-
tion factors, thus providing a direct link between signalling
molecules that control these processes and transcriptional
responses.
(1)
The basic concept of a NR signalling pathway
involves production of the signal (endocrine, paracrine,
autocrine or even intracrine), its transport to peripheral
organs, binding of the ligand to the receptor, and transcrip-
tional activation of the receptor.
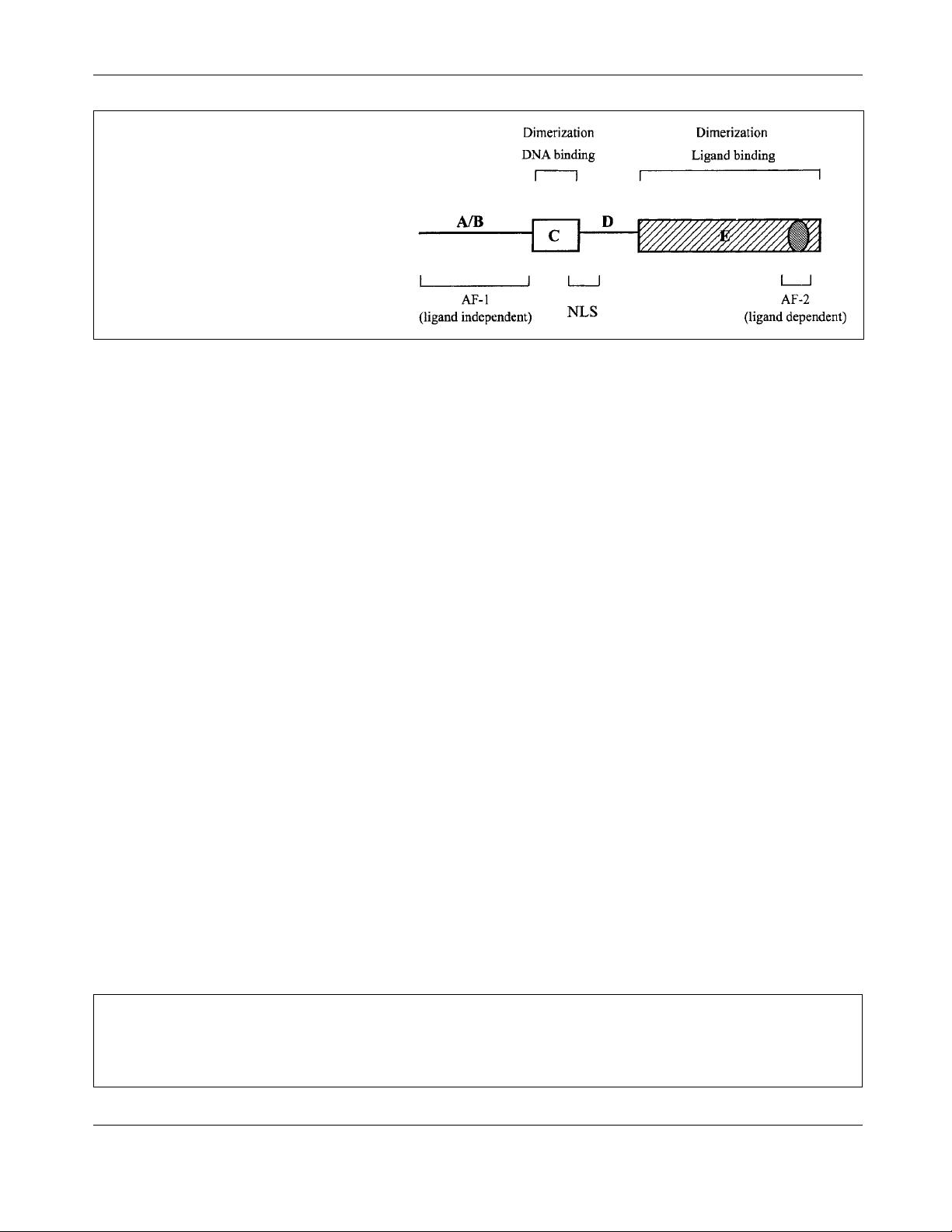
NRs share a common structural organization with a
central, well-conserved DNA-binding domain (DBD, also
termed C domain), a variable N-terminal region (A/B domain)
a non-conserved hinge (D domain) and a carboxy-terminus,
moderately conserved ligand-binding domain (LBD, E
domain) (Fig. 1).
NRs are phylogenetically related proteins clustered into a
large superfamily, including receptors for hydrophobic
molecules such as steroid hormones (estrogens, glucocorti-
coids, progesterone, mineralocorticoids, androgens, vitamin
D, ecdysone, oxysterols, bile acids etc), retinoic acids (all-
trans and 9-cis isoforms), thyroid hormones, fatty acids,
leukotrienes and protaglandins (Fig. 2). In addition, a number
of receptors, for which ligands either do not exist or still have
to be identified have been described and are referred to as
orphan receptors. NRs are promising pharmacological
targets as they bind small molecules, which can be easily
modified by drug design, and control functions associated
with major pathologies (cancer, osteoporosis, diabetes, etc).
The development of synthetic ligands for ER, RAR and
PPARg has, for instance, already led to pharmacological
applications for cancer and type 2 diabetes. Because of
these important therapeutic applications, the search of
ligands for orphan receptors and the identification of novel
signalling pathways has become a very active research
field.
(2)
The structural diversity of the ligands contrasts with the
conservation and mode of action of their receptors. This, and
the large number of orphan receptors, have prompted much
speculation on the origins of this signalling pathway. A
classical view suggests that orphan receptors have evolved
as liganded molecules, which through gene duplication
reached the present day diversity.
(3)
According to this model,
orphan receptors would have lost their ability to bind ligands
recently in evolution. We recently suggested an alternative
hypothesis, in which ligand binding was acquired during NRs
evolution.
(4)
The ancestral receptor would have been an
orphan receptor and the various liganded receptors have
gained ligand recognition independently during evolution. In
BioEssays 22:717±727, ß 2000 John Wiley & Sons, Inc. BioEssays 22.8 717
Ecole Normale Supe
Â
rieure de Lyon, Lyon, France.
Funding agencies: EMBO, Fondation pour la Recherche Me
Â
dicale,
ARC, LNCC, CNRS and MENRT.
*Correspondence to: Vincent Laudet, UMR 5665 du CNRS, Ecole
Normale Supe
Â
rieure de Lyon, 46 alle
Â
e d'Italie, 69364 Lyon Cedex 07,
France. E-mail: Vincent.Laudet@ens-lyon.fr
Abbreviations: AF2-AD, activation function 2-autonomous domain,
AR, androgen receptor, CARb, constitutive activated receptor b, DBD,
DNA-binding domain, DR, direct repeat, EcR, ecdysone receptor, ER,
estrogen receptor, ERKO, estrogen receptor knock out, ERR,
estrogen related receptor, FXR, farnesol X receptor, GCNF, germ
cell nuclear factor, GR, glucocorticoid receptor, HRE, hormone
response element, LBD, ligand-binding domain, LXR, liver X receptor,
MR, mineralocorticoid receptor, NGFIB, nerve growth factor induced
gene B, NR, nuclear receptor, PPAR, peroxisome proliferating
activated receptor, PR, progesterone receptor, PRKO, progesterone
receptor knock out, PXR, pregnane X receptor, RAR, retinoic acid
receptor, ROR, retinoic and related orphan receptor, RXR, retinoic X
receptor, SF1, steroidogenic factor 1, SRC-1, steroid receptor
coregulator 1, T3, triiodothyronine, TR, thyroid hormone receptor,
USP, ultraspiracle, VDR, vitamin D receptor.
Review articles
this review, we discuss arguments from phylogenetic,
functional and structural studies that support a recent gain
of ligand-binding ability in classical receptors, rather than a
loss of ligand binding in orphan receptors during metazoan
evolution.
How to turn on a NR?
NRs bind as homodimers or heterodimers to specific DNA
sequences, termed hormone response elements (HRE),
which are most often formed by a repetition of the core
consensus motif PuGGTCA.
(1)
For example, steroid recep-
tors bind as homodimers to HREs containing two core motifs
separated by 1±3 nucleotides and organised as palin-
dromes. A number of receptors, including retinoic acid
receptors (RARs), thyroid hormone receptors (TRs), and
some orphan receptors (NGFIB), heterodimerize with re-
tinoid X receptors (RXRs) and bind to direct repeats (DR) of
the core motif. The spacing between the two halves of the DR
dictates the type of heterodimer that will bind.
(1)
For example,
a DR separated by five nucleotides (DR5) will be most often
recognised by an RXR-RAR heterodimer, whereas a DR4
will be bound by an RXR-TR heterodimer. Some receptors,
such as the SF1, ROR or Rev-erb orphan receptors, can
bind to DNA as monomers through a single core sequence.
In this case, an A/T-rich region 5
0
to the core element
governs the binding specificity. At present, only orphan
receptors are known to bind as monomers to these half sites.
It was recently shown that some orphan receptors (e.g. ERR-
1, GCNF) can also bind a single core motif as dimers.
(5,6)
Many NRs are transcriptional silencers in the absence of
ligand (apo-receptor) as a result of interaction with inter-
mediary factors known as co-repressors.
(7,8)
Determination
of the three-dimensional structure of NRs has shown that the
LBD region is structured as a three-layered a-helical anti-
parallel sandwich of 12 helices forming an hydrophobic
pocket. Upon ligand binding (holo-receptor), the ligand
makes different contacts with amino-acid residues, promot-
ing a conformational change that closes the ``lid'' (helix 12,
H12) on the pocket. Thus the activation domain within the
H12 (AF2-AD) is able to interact with co-activators and
promotes transcription of target genes.
(9±12)
The conforma-
tional change of the LBD upon ligand binding is therefore
necessary for the transactivation function of NRs. It is
believed that co-repressors and co-activators modulate
transcription at least in part by modifying the status of
histone acetylation in chromatin. An opposite mode of action
was demonstrated for the orphan receptor CARb, for which
binding of steroid-like compounds (androstanol) was shown
to repress its transcriptional activity.
(13)
In the apo-conforma-
tion, CARb interacts with co-activators such as SRC-1,
whereas androstanol binding reverses this interaction.
In summary, a NR ligand can be defined as a small
hydrophobic molecule that directly binds to the LBD and
promotes a structural rearrangement allowing the AF2-AD
region to interact with transcriptional coregulators (co-
activators, co-repressors) that will connect the NR to the
basic transcriptional machinery, resulting in the transcrip-
tional stimulation/silencing of target genes.
Evolution of NRs
Alignment of the well-conserved DBD and LBD, and
phylogenetic analysis has resulted in an evolutionary tree
Figure 1. Structure of an archetypal nuclear hor-
mone receptor. The two conserved domains, C and
E, are boxed and their main functions are indicated.
AF-1 and AF-2AD, located, respectively, in N and C
termini are the trans-activation domains. NLS is the
nuclear localization signal.
Figure 2. Chemical structure of endogenous known nuclear receptor ligands and activators. Their respective K
d
or EC
50
are indicated.
Binding/activation by palmitoyl-CoA and 25-hydroxycholesterol to/of HNF4 and SF1 receptors respectively have not been reproduced.
The name of each group (i. e. subfamily name) according to the recent nomenclature
(18)
is indicated below the chemical name (see
Fig. 3).
Review articles
718 BioEssays 22.8
剩余10页未读,继续阅读
资源评论

weixin_38613173
- 粉丝: 3
- 资源: 928
上传资源 快速赚钱
 我的内容管理
展开
我的内容管理
展开
 我的资源
快来上传第一个资源
我的资源
快来上传第一个资源
 我的收益 登录查看自己的收益
我的收益 登录查看自己的收益 我的积分
登录查看自己的积分
我的积分
登录查看自己的积分
 我的C币
登录后查看C币余额
我的C币
登录后查看C币余额
 我的收藏
我的收藏  我的下载
我的下载  下载帮助
下载帮助

 前往需求广场,查看用户热搜
前往需求广场,查看用户热搜最新资源
- 基于matlab的SVM二分类代码,可替数据集直接使用,算法包括数据归一化处理、c,g参数寻优,生成c,g最佳参数等高线图,交叉准确率与cg关系图及SVM预测结果图
- MATLAB制动能量回收模型simulink 轮毂电机再生制动模型 MATLAB液压电机制动模型 电动车再生制动模型 截图说明,简单易懂,一一对应 电动汽车再生制动控制策略模型,逻辑门限值控制算法
- 自动泊车APA项目开发,C代码开发泊车路径规划
- MMC型APF,MMC型statcom,MMC型储能系统,MMC-HVDC,MMC型整流器,逆变器,MMC,模块化多电平变器,MMCsimulink仿真,最近电平逼近调制,载波移相调制,排序法均衡
- a*算法与人工场势法结合的算法
- 非隔离双向DC DC变器 buck-boost变器仿真 输入侧为直流电压源,输出侧接蓄电池 模型采用电压外环电流内环的双闭环控制方式 正向运行时电压源给电池恒流恒压充电,反向运行时电池放电维持直流侧电
- 永磁同步模型电流预测控制 电流环采用预测控制双矢量改进算法 含有相关学习文献
- 行业领先的永磁同步电机pmsm 无感foc方案评估套件 套件包括电机与电路板 1、新工程采用闭环结构速度位置观测器,相比较上一代观测器; 1)低速性能进一步提升,新方案在6010电机5hz带载
- 基于Crowbar电路的双馈风力发电机DFIG低电压穿越LVRT仿真模型 Matlab Simulink仿真模型(成品) 本模型采用Crowbar保护电路(串电阻)实现低电压穿越,在电网电压跌落时投入
- 三相异步电机直接转矩DTC控制 Matlab Simulink仿真模型(成品) 传统策略DTC 1.转速环采用PI控制 2.转矩环和磁链环采用滞环控制 3.含扇区判断、磁链观测、转矩控制、开关状态选择
- 基于莱维飞行格和随机游动策略的灰狼优化算法 Matlab 源码 改进点: 1. 分段可调节衰减因子 2. 莱维飞行和随机游动策略 3. 贪婪算法寻优
- matlab simulink 在自带系统的单相MMC模型上实现了最近电平逼近调制,模型和函数是自己改的
- 松下FP系列程序 松下FP系列程序,搭配松下伺服,昆仑通态触摸屏锂电池全自动叠片贴胶机 大型程序近30000步,三个PLC,主从站通信控制 ,隔膜放卷纠偏控制,正负极真空取料叠片控制,可设定叠片
- 该模型在额定以下采用MTPA控制,速度环输出给定电流,然后代入MTPA得到dq电流,电压反馈环输出超前角进行弱磁 PI控制采用抗积分饱和,SVPWM考虑过调制情况,附带参考文献
- 自动驾驶代码-Ros移植Apollo规划方案,可编译运行,包含autoware的Lanelet2框架 帮助大家快速入门实践 完善代码,加功能等
- 一阶RC模型自适应遗忘因子递推最小二乘法+扩展卡尔曼滤波算法AFFRLS+EKF锂电池参数和SOC联合估计 遗忘因子可随时间自适应变化,不再是定值,提高估计精度 matlab程序 参考文献
资源上传下载、课程学习等过程中有任何疑问或建议,欢迎提出宝贵意见哦~我们会及时处理!
点击此处反馈



安全验证
文档复制为VIP权益,开通VIP直接复制
 信息提交成功
信息提交成功