IEEE
Proof
IEEE TRANSACTIONS ON NANOBIOSCIENCE, VOL. 14, NO. 5, JULY 2015 1
An Efficient Algorithm for Discovering Motifs in
Large DNA Data Sets
Qiang Yu, Hongwei Huo*, Member, IEEE, Xiaoyang Chen, Haitao Guo, Jeffrey Scott Vitter, Fellow, IEEE,
and Jun Huan, Member, IEEE
Abstract—The planted motif discovery has been success-
fully used to locate transcription factor binding sites in dozens
of promoter sequences over the past decade. However, there h
as
not been enough work done in identifying
motifs in the
next-generation sequencing (ChIP-seq) data sets, which contain
thousands of input sequences and thereby bring new
challenge
to make a good identification in reasonable time. To cater this
need, we propose a new planted
motif discovery algorithm
named MCES, which identifies motifs by mining and
combining
emerging substrings. Specially, to handle larger data sets, we
designaMapReduce-basedstrategytomineemergingsubstrings
distributedly. Experimental results on
the simulated data show
that i) MCES is able to identify
motifs efficiently and effec-
tively in thousands to millions of input sequences, and runs faster
than the state-of-the-art
motif
discovery algorithms, such as
F-motif and TraverStringsR; ii) MCES is able to identify motifs
without known lengths, and has a better identification accuracy
than the competing algorithm CisFin
der. Also, the validity of
MCES is tested on real data sets. MCES is freely available at
http://sites.google.com/site/feqond/mces.
Index Terms—ChIP-seq, emerging substrings, MapReduce,
motif discovery.
I. INTRODUCTION
M
OTIF discovery is an important and challenging
problem in computationa
l biology. It plays a key role
in locating transcription factor binding sites (TFBS) in DNA
sequences. Binding sites tend to be short and degenerate, so it
is difficult to disting
uish them from the input sequences. The
planted
motif discovery [1] is a famous formulation for
motif discovery, which has been proven to be NP-complete [2].
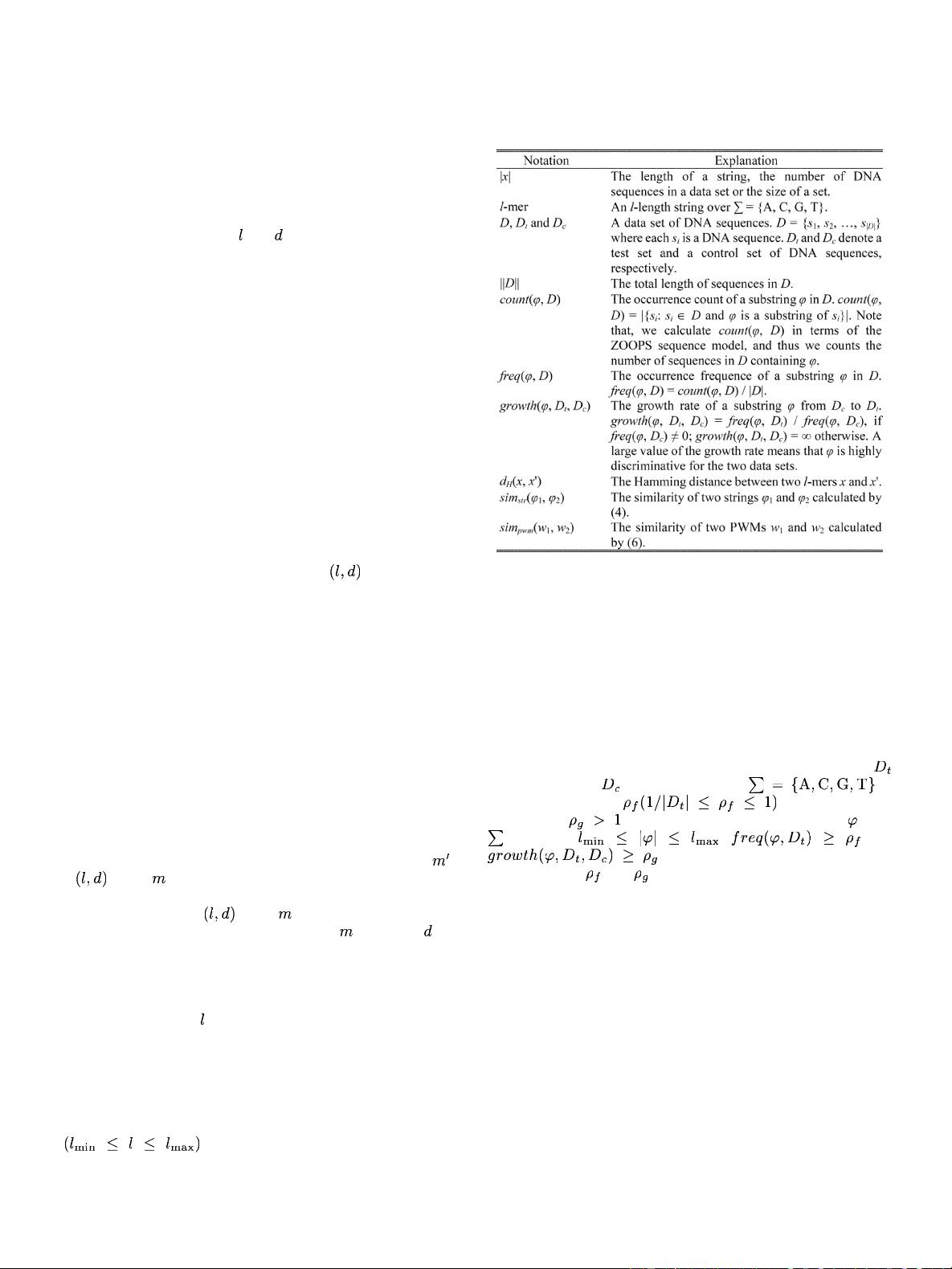
Planted
Moti
f Discovery Problem: Given a set of
-length DNA sequences over the alphabet
and two nonnegative integers and , satisfying
,thetas
k is to find one or more
-length strings
such that occurs in all or a large fraction of the sequences
with up to
mismatches. The -length string is called an
Manuscript received March 27, 2015; accepted March 31, 2015. Asterisk in-
dicates corresponding author.
Q. Yu, X. Chen, and H. Guo are with the School of Computer Science and
Technology, Xidian University, Xi'an, 710071, China (e-mail: qyu@mail.xi-
dian.edu.cn; xychen@mail.xidian.edu.cn; htguo@mail.xidian.edu.cn).
*H. Huo is with the School of Computer Science and Technology, Xidian
University, Xi'an, 710071, China (e-mail: hwhuo@mail.xidian.edu.cn).
J. S. Vitter and J. Huan are with the Information and Telecommunication of
Technology Center, The University of Kansas, Lawrence, 66047, USA (e-mail:
{jsv,jhuan}@ku.edu).
This work was supported in part by the National Natural Science Foundation
of China under Grant 61173025 and 61373044, and the Fundamental Research
Funds for the Central Universities under Grant JB150306 and XJS15014.
Digital Object Identifier 10.1109/TNB.2015.2421340
motif and each occurrence of is called a motif instance
of
.
According to how and where motif occurrences appear in the
sequences, there are three types of motif discovery sequence
model: OOPS, ZOOPS and TCM [3], corresponding to one
oc-
currence per sequence, zero or one occurrence per sequence
and zero or more occurrences per sequence, respectively. The
ZOOPS and TCM sequence model are more consistent
with the
real biological situation than the OOPS model, but identifying
motifs under these two models is more difficult than that under
the OOPS model.
Numerous algorithms have been proposed to identify motifs
in several to dozens of promoter sequences from co-regulated
or homologous genes [4]. These algorit
hms can be divided
into two categories in terms of the used motif representation
models: those using consensus sequences [5] and those using
position weight matrices (PWM) [6].
Most identification al-
gorithms based on consensus sequences are pattern-driven
[7]–[11]. They traverse all sequence patterns of length
with
an initial search space of
and report all motifs.
The identification algorithms based on PWM usually employ
statistical techniques [3], [12]. They iteratively update an initial
PWM and report the motif with
high score.
In recent years, the novel experimental techniques, such
as protein-binding microarray (PBM) [13] and chromatin
immunoprecipitation f
ollowed by high-throughput sequencing
(ChIP-seq) allows the genome-wide identification of TFBSs
[4], [14]. The experiments can output a list of transcription
factor binding regio
ns (i.e., peak regions), but motif discovery
methods are still needed to accurately locating TFBSs in these
peak regions. The advantage of a ChIP-seq data set is that the
sequences are cle
aner than the traditional promoter sequences
[4]. That is, not only a high percentage of sequences contain
TFBSs, but also each sequence has a high resolution (i.e., the
sequence length
is short, about 200 base pairs). It seems easier
for motif discovery methods to obtain a high identification ac-
curacy in ChIP-seq data sets, but the size of a ChIP-seq data set
is very large a
nd the set contains thousands or more sequences,
requiring a high computational efficiency of motif discovery.
Unfortunately, almost all algorithms designed for identifying
motifs in pr
omoter sequences, either the pattern-driven algo-
rithms or statistical algorithms, become too time-consuming
for ChIP-seq data sets.
ChIP-tai
lored versions of traditional motif discovery algo-
rithms have been proposed, such as MEME-ChIP [15]. These
algorithms usually present limitations on the data set size by se-
lectin
g a small subset of the sequences to make motif identifica-
tion [16]. For example, MEME-ChIP just selects 600 sequences
at random from the input sequences and then identifies motifs
by usi
ng the expectation-maximization algorithm. In spite of
this, these algorithms still show a poor time performance due
1536-1241 © 2015 IEEE. Personal use is permitted, but republication/redistribution requires IEEE permission.
See http://www.ieee.org/publications_standards/publications/rights/index.html for more information.


 我的内容管理
展开
我的内容管理
展开
 我的资源
快来上传第一个资源
我的资源
快来上传第一个资源
 我的收益 登录查看自己的收益
我的收益 登录查看自己的收益 我的积分
登录查看自己的积分
我的积分
登录查看自己的积分
 我的C币
登录后查看C币余额
我的C币
登录后查看C币余额
 我的收藏
我的收藏  我的下载
我的下载  下载帮助
下载帮助 
 前往需求广场,查看用户热搜
前往需求广场,查看用户热搜

 信息提交成功
信息提交成功