The GPML Toolbox version 4.2
Carl Edward Rasmussen & Hannes Nickisch
August 22, 2018
Abstract
The GPML toolbox is an Octave 3.2.x and Matlab 7.x implementation of inference and pre-
diction in Gaussian process (GP) models. It implements algorithms discussed in Rasmussen &
Williams: Gaussian Processes for Machine Learning , the MIT press, 2006 and Nickisch &
Rasmussen: Approximations for Binary Gaussian Process Classification , JMLR, 2008.
The strength of the function lies in its flexibility, simplicity and extensibility. The function is
flexible as firstly it allows specification of the properties of the GP through definition of mean func-
tion and covariance functions. Secondly, it allows specification of different inference procedures,
such as e.g. exact inference and Expectation Propagation (EP). Thirdly it allows specification of
likelihood functions e.g. Gaussian or Laplace (for regression) and e.g. cumulative Logistic (for
classification). Simplicity is achieved through a single function and compact code. Extensibility is
ensured by modular design allowing for easy addition of extension for the already fairly extensive
libraries for inference methods, mean functions, covariance functions and likelihood functions.
This document is a technical manual for a developer containing many details. If you are not
yet familiar with the GPML toolbox, the user documentation and examples therein are a better
way to get started.
1

Contents
1 Gaussian Process Training and Prediction 3
2 The gp Function 4
3 Inference Methods 9
3.1 Exact Inference with Gaussian likelihood . . . . . . . . . . . . . . . . . . . . . . . . 10
3.2 Laplace’s Approximation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11
3.3 Expectation Propagation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11
3.4 Kullback Leibler Divergence Minimisation . . . . . . . . . . . . . . . . . . . . . . . . 12
3.5 Variational Bayes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 13
3.6 Compatibility Between Inference Methods and Covariance Approximations . . . . . . 13
3.7 Sparse Covariance Approximations . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
3.8 Grid-Based Covariance Approximations . . . . . . . . . . . . . . . . . . . . . . . . . 14
3.9 State Space Representation of GPs . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
4 Likelihood Functions 18
4.1 Prediction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18
4.2 Interface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
4.3 Implemented Likelihood Functions . . . . . . . . . . . . . . . . . . . . . . . . . . . . 21
4.4 Usage of Implemented Likelihood Functions . . . . . . . . . . . . . . . . . . . . . . . 22
4.5 Compatibility Between Likelihoods and Inference Methods . . . . . . . . . . . . . . . 23
4.6 Gaussian Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23
4.6.1 Exact Inference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
4.6.2 Laplace’s Approximation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
4.6.3 Expectation Propagation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
4.6.4 Variational Bayes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
4.7 Warped Gaussian Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
4.8 Gumbel Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
4.9 Laplace Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
4.10 Student’s t Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 30
4.11 Cumulative Logistic Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31
4.12 GLM Likelihoods: Poisson, Negative Binomial, Weibull, Gamma, Exponential, In-
verse Gaussian and Beta . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 32
4.12.1 Inverse Link Functions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 32
4.12.2 Poisson Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34
4.12.3 Weibull Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34
4.12.4 Gamma Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34
4.12.5 Exponential Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
4.12.6 Inverse Gaussian Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
4.12.7 Beta Likelihood . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
5 Mean Functions 36
5.1 Interface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
5.2 Implemented Mean Functions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37
5.3 Usage of Implemented Mean Functions . . . . . . . . . . . . . . . . . . . . . . . . . . 37
6 Covariance Functions 39
6.1 Interface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 39
6.2 Implemented Covariance Functions . . . . . . . . . . . . . . . . . . . . . . . . . . . . 43
6.3 Usage of Implemented Covariance Functions . . . . . . . . . . . . . . . . . . . . . . . 44
2

7 Hyperpriors 46
7.1 Interface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 46
7.2 Implemented Hyperpriors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 48
7.3 Usage of Implemented Hyperpriors . . . . . . . . . . . . . . . . . . . . . . . . . . . . 48
3
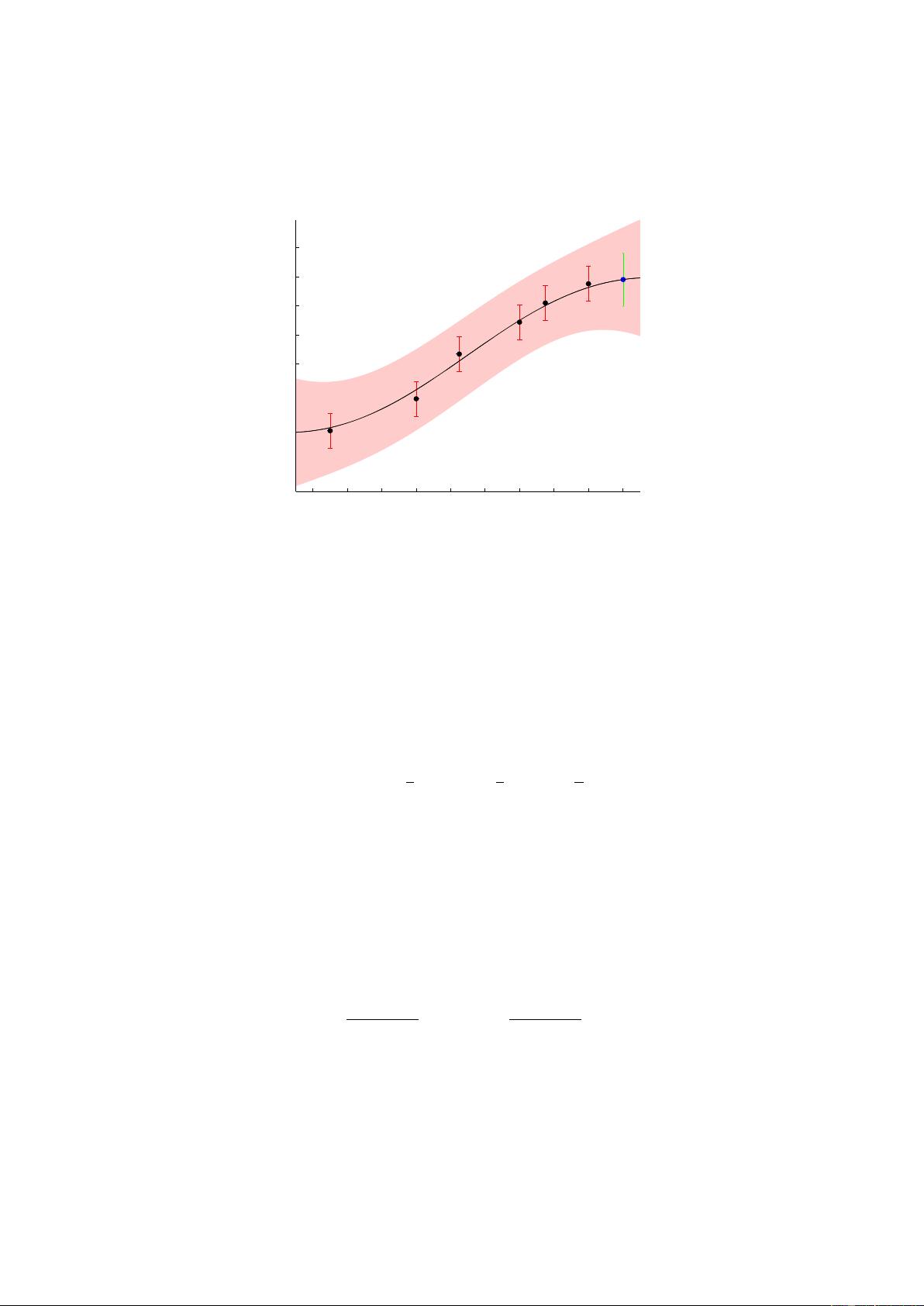
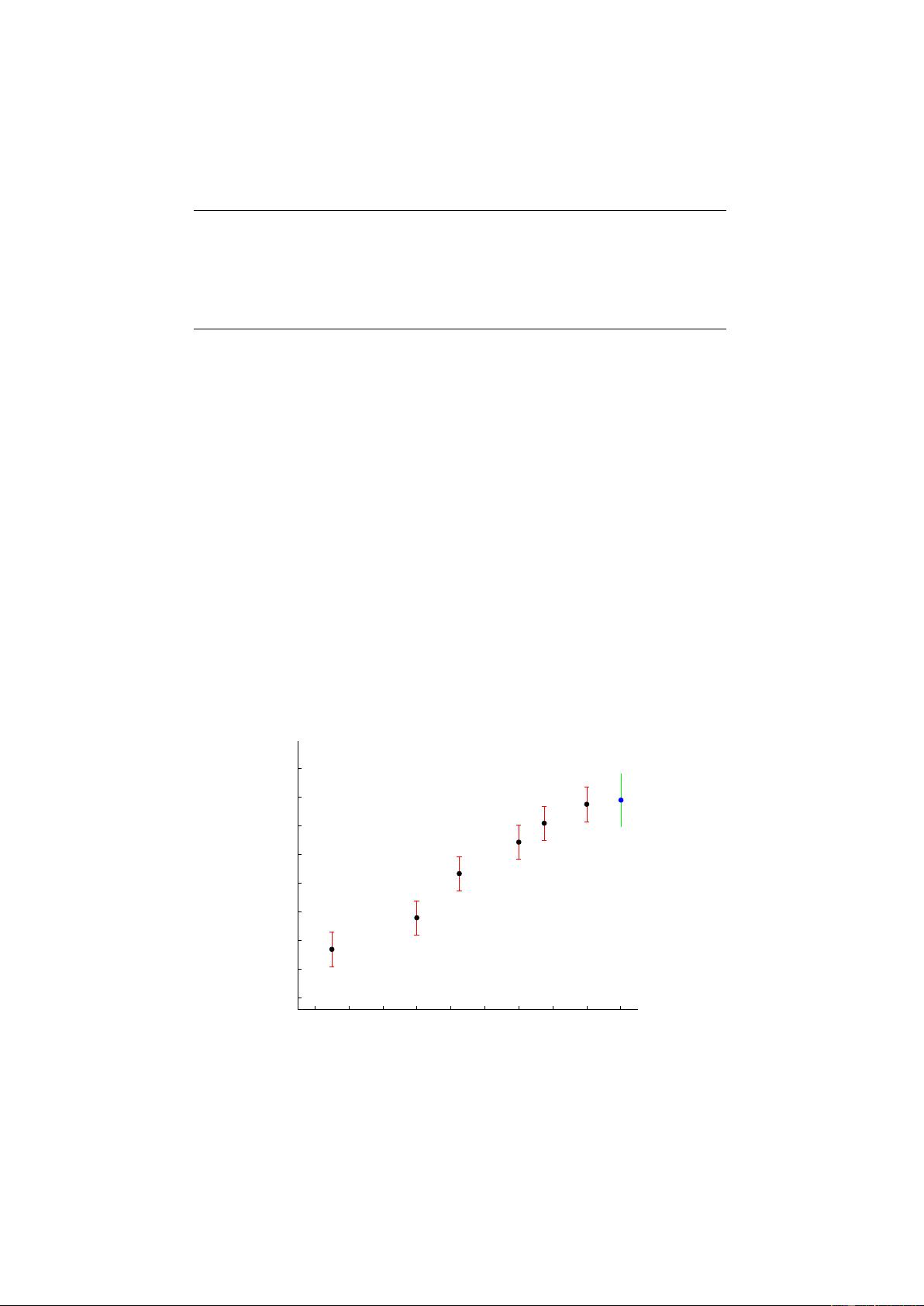
1 Gaussian Process Training and Prediction
The gpml toolbox contains a single user function gp described in section 2. In addition there are a
number of supporting structures and functions which the user needs to know about, as well as an
internal convention for representing the posterior distribution, which may not be of direct interest to
the casual user.
Inference Methods: An inference method is a function which computes the (approximate) poste-
rior, the (approximate) negative log marginal likelihood and its partial derivatives w.r.t.. the
hyperparameters, given a model specification (i.e., GP mean and covariance functions and a
likelihood function) and a data set. Inference methods are discussed in section 3. New in-
ference methods require a function providing the desired inference functionality and possibly
extra functionality in the likelihood functions applicable.
Hyperparameters: The hyperparameters is a struct controlling the properties of the model, i.e.. the
GP mean and covariance function and the likelihood function. The hyperparameters is a struct
with the three fields mean, cov and lik, each of which is a vector. The number of elements in
each field must agree with number of hyperparameters in the specification of the three functions
they control (below). If a field is either empty or non-existent it represents zero hyperparam-
eters. When working with FITC approximate inference, the inducing inputs xu can also be
treated as hyperparameters for some common stationary covariances.
Hyperparameter Prior Distributions: When optimising the marginal likelihood w.r.t. hyperparame-
ters, it is sometimes useful to softly constrain the hyperparameters by means of prior knowl-
edge. A prior is a probability distribution over individual or a group of hyperparameters,
section 7.
Likelihood Functions: The likelihood function specifies the form of the likelihood of the GP model
and computes terms needed for prediction and inference. For inference, the required properties
of the likelihood depend on the inference method, including properties necessary for hyperpa-
rameter learning, section 4.
Mean Functions: The mean function is a cell array specifying the GP mean. It computes the mean
and its derivatives w.r.t.. the part of the hyperparameters pertaining to the mean. The cell array
allows flexible specification and composition of mean functions, discussed in section 5. The
default is the zero function.
Covariance Functions: The covariance function is a cell array specifying the GP covariance function.
It computes the covariance and its derivatives w.r.t.. the part of the hyperparameters pertaining
to the covariance function. The cell array allows flexible specification and composition of
covariance functions, discussed in section 6.
Inference methods, see section 3, compute (among other things) an approximation to the posterior
distribution of the latent variables f
i
associated with the training cases, i = 1, . . . , n. This approx-
imate posterior is assumed to be Gaussian, and is communicated via a struct post with the fields
post.alpha, post.sW and post.L. Often, starting from the Gaussian prior p(f) = N(f|m, K) the
approximate posterior admits the form
q(f|D) = N
f|µ = m + Kα, V = (K
−1
+ W)
−1
, where W diagonal with W
ii
= s
2
i
. (1)
In such cases, the entire posterior can be computed from the two vectors post.alpha and post.sW;
the inference method may optionally also return L = chol(diag(s)K diag(s) + I).
If on the other hand the posterior doesn’t admit the above form, then post.L returns the matrix
L = −(K + W
−1
)
−1
(and post.sW is unused). In addition, a sparse representation of the posterior
may be used, in which case the non-zero elements of the post.alpha vector indicate the active entries.
4

2 The gp Function
The gp function is typically the only function the user would directly call.
4a hgp.m 4ai≡
1 function [varargout] = gp(hyp, inf, mean, cov, lik, x, y, xs, ys)
2 hgp function help 4bi
3 hinitializations 5bi
4 hinference 6ci
5 if nargin==7 % if no test cases are provided
6 varargout = {nlZ, dnlZ , post}; % report -log marg lik, derivatives and post
7 else
8 hcompute test predictions 7i
9 end
It offers facilities for training the hyperparameters of a GP model as well as predictions at unseen
inputs as detailed in the following help.
4b hgp function help 4bi≡ (4a)
1 % Gaussian Process inference and prediction. The gp function provides a
2 % flexible framework for Bayesian inference and prediction with Gaussian
3 % processes for scalar t argets, i.e. both regression and binary
4 % classification. The prior is Gaussian process , defined through specification
5 % of its mean and covariance function. The likelihood function i s also
6 % specified. Both the prior and the likelihood may have hyperparameters
7 % associated with them.
8 %
9 % Two modes are possible: training or prediction: if no test cases are
10 % supplied, then the negative log marginal likelihood and its partial
11 % derivatives w.r.t. the hyperparameters is computed; this mode is u sed to fit
12 % the hyperparameters. If test c ases are given, then the test set predictive
13 % probabilities are returned. Usage:
14 %
15 % training: [nlZ dnlZ ] = gp(hy p, inf, mea n , cov, lik, x, y);
16 % prediction: [ymu ys2 fmu fs2 ] = gp(hyp, inf, mean, cov, lik, x, y, xs);
17 % or: [ymu ys2 fmu fs2 lp] = gp(hyp, inf, mean, cov, lik, x, y, xs, ys);
18 %
19 % where:
20 %
21 % hyp struct of column vectors of mean/cov/lik hyperparameters
22 % inf function specifying the inference method
23 % mean prior mean function
24 % cov prior covariance function
25 % lik likelihood function
26 % x n by D matrix of training inputs
27 % y column vector of length n of training targets
28 % xs ns by D matrix of test inputs
29 % ys column vector of length nn of test targets
30 %
31 % nlZ returned value of the negative log marginal likelihood
32 % dnlZ struct of column vectors of partial derivatives of the negative
33 % log marginal likelihood w.r.t. mean/cov/lik hyperparameters
34 % ymu column vector (of length ns) of predictive output means
35 % ys2 column vector (of length ns) of predictive output variances
36 % fmu column vector (of length ns) of predictive latent means
37 % fs2 column vector (of length ns) of predictive latent variances
38 % lp column vector (of l ength ns) of log predictive probabilities
39 %
5

















