in some dictionary, which may be identified based on the properties
of signals at hand. Recently, dictionaries learnt from the data were
found to have potential for several applications. Several interesting
dictionary learning methods like K-SVD [8] and Method of Optimal
Directions (MOD) [13] were developed to provide each member of
database with sparse representation. The emerging filed of com-
pressed sensing has a potential for exploiting sparsity present in
medical images. This work is an attempt towards proposing a new
CBMIR technique that relies on sparsity based concepts.
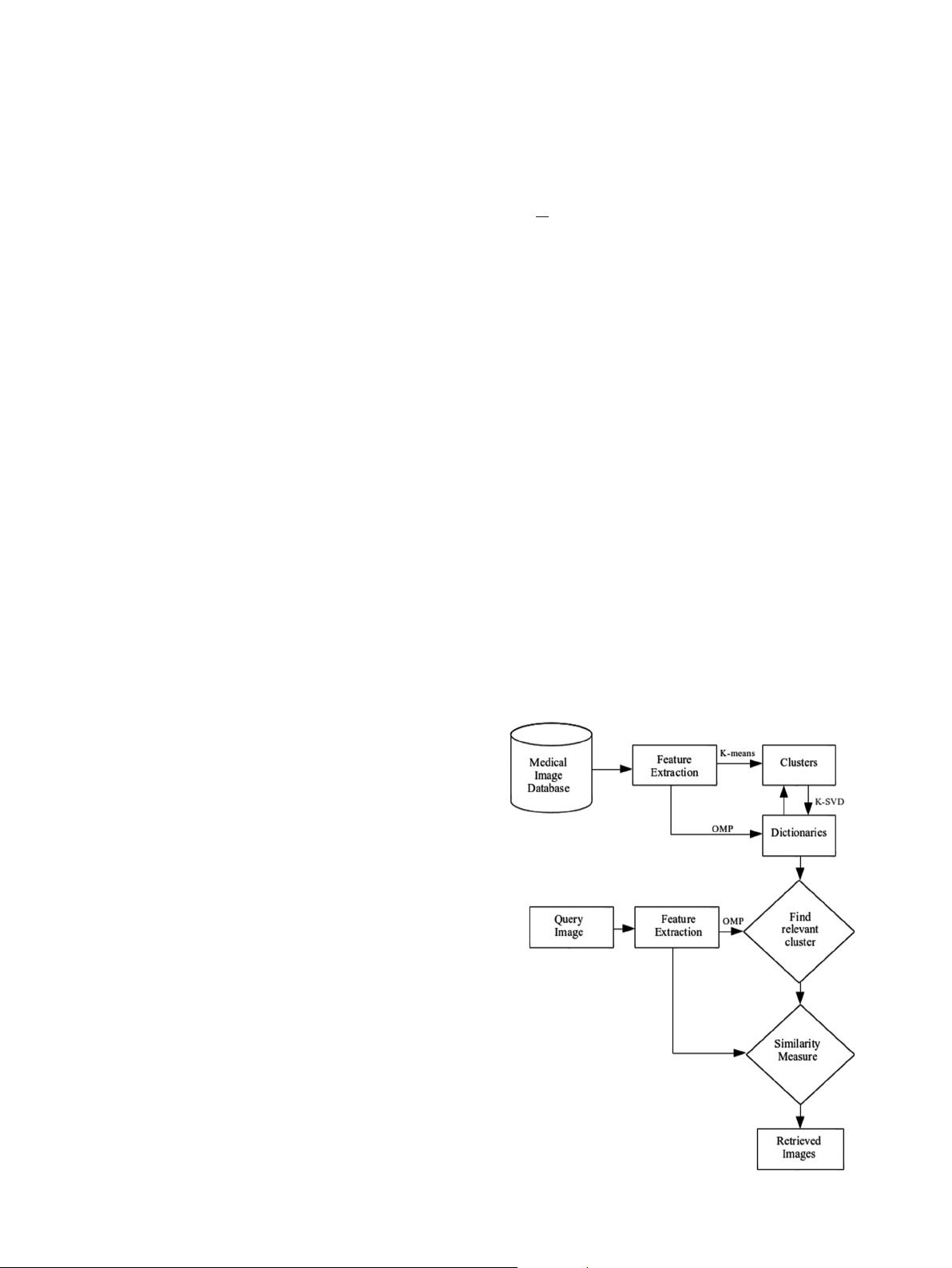
In particular, we propose a dictionary based clustering algorithm
for grouping the images in medical databases. This clustering
technique increases the retrieval speed and improves the accuracy
of the results. The dictionary based methods rely on the premise
that two signals belonging to the same cluster have decomposition
in terms of similar atoms (columns) of a dictionary. Making use of
this property, we match the input query with the appropriate
cluster. The selection of features for adequately representing the
class specific information is an important step in CBIR. For this, we
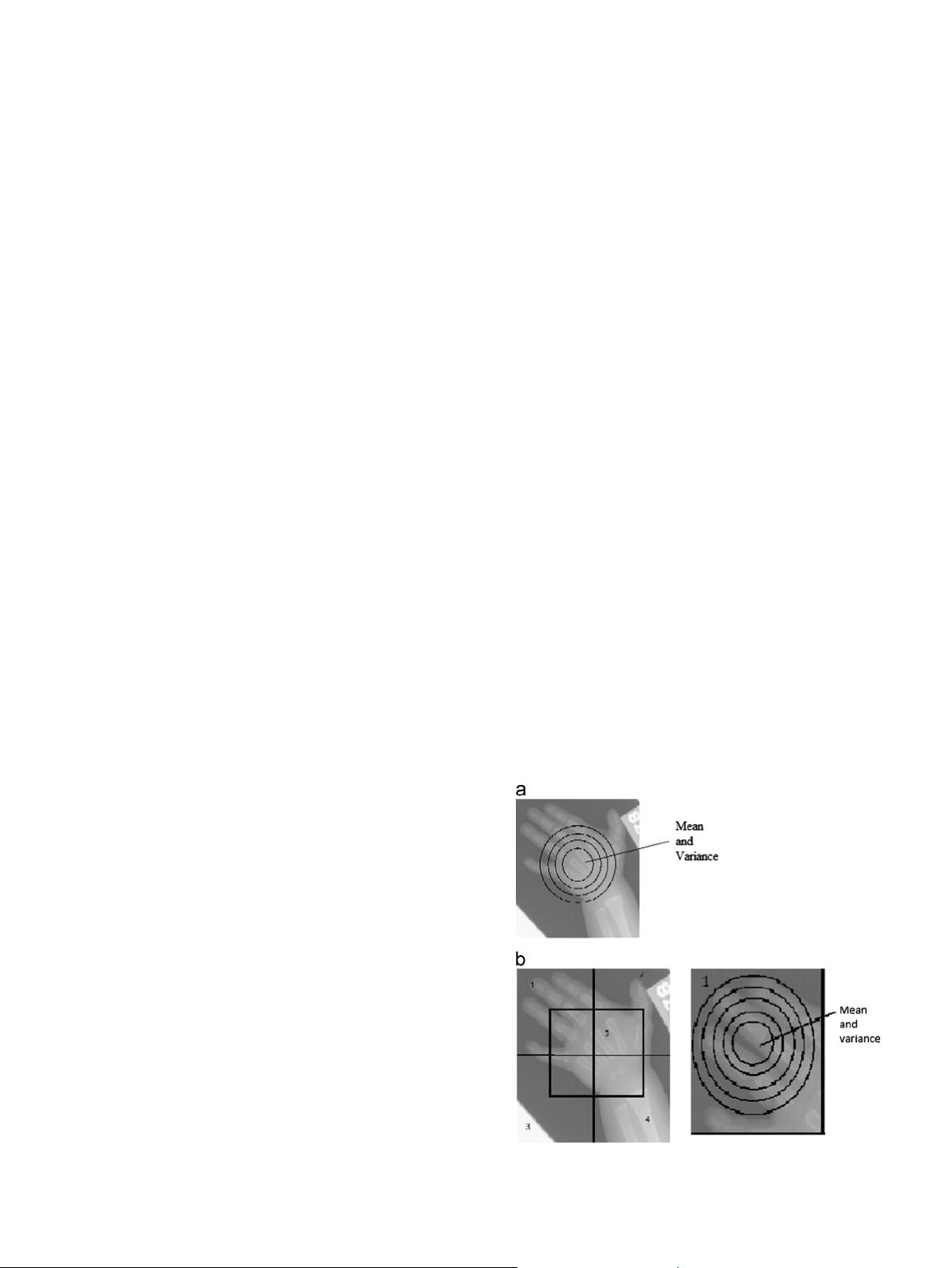
divide the image into four sub-images of equal size. In addition, we
consider another sub-image which is of same size as other four sub-
images to capture the rich information available at the center of
medical images. We then partition each sub-image into concentric
circular regions around the center, and consider the mean and
variance of pixel intensities in each region as components in the
feature vector. Some image retrieval methods were proposed in the
literature which made use of SVM [9–11].Itistobeemphasized
here that K-SVD and SVM based methods are different in the sense
that K-SVD is a dictionary learning approach banking on the
concept of sparsity, which is not the case with SVM. While SVM
requires some training data, the way we use K-SVD in the present
work does not require any labeled data. The present CBMIR
technique centers around images produced in radiology. As color
and shape features are of less importance in medical domain [3],we
use texture features in the present work.
The work done in this paper has the objective of categorizing (and
retrieving) radiological images consisting of differ ent organs, mod-
ality , views. W e demonstrate the usefulness of our approach through
exte nsive experimental results. For a given N, the number of clusters,
we design N dictionaries to represent the clusters. W e associate an
image of database to a dictionary based on the sparsity crit erion.
Given a query image, we invoke the concept of sparsity to identify
appropriate cluster, wherein we sear ch for relev ant images. The rest of
the paper is organized as follows: Sections 2 and 3 give brief accounts
of a survey of related works and dictionary learning. Section 4
presents the proposed content based medical image retriev al using
dictionary learning method. Experiments of CBMIR application are
discussed in detail in Section 5.Finally,Section6 concludesthispaper.
2. Related work
Chu et al. [16] described a knowledge based image retrieval of
computed tomography (CT) and magnetic resonance imaging (MRI)
images. In this approach, the brain lesions were automatically
segmented and represented through a knowledge based semantic
model. Cai et al. [17] proposed a CBIR system for functional dynamic
positron emission tomography (PET) images of the human brain,
where clusters of tissue time activity from the temporal domain
were used in the computation of similarity measure for retrieval. In
[18], the delineations of the regions of interest were manually
performed on the key frame from the stack of high resolution CT
images. These were used as features to represent the entire image.
In the Bag-Of-W ords (BO W) [5] framework, the image patches
were sampled densely or sparsely by “interest points”
detectors and
were depicted by local patch descriptors like SIFT . These descriptors
were used to classify liver lesions in CT images. In [6], a te xtur e based
analysis of lung CT images was proposed through Riesz w avelets.
This method used SVM to learn the respective relevance of multi-
scale components. Guimond et al. [1 9] introduced user -select ed
volume of interest (VOI) for the retriev al of pathologic al brain MRI
images. In [2 1], group sparse representation with dictionary learning
for medical image denoising and fusion was used. W a velet optimiza-
tion techniques for content based image retriev al in medical database
were described in Quellec et al. [22]. Linear discriminate analysis
(LD A) based selection and feature extraction algorithm for classifica-
tion and segmentation of one dimensional radar signals and two-
dimensional te xtur e and document images using w a velet packet was
proposed by Etemand and Chellappa [23]. Recently , similar algo-
rithms for simultaneous sparse signal representation and discrimina-
tion were proposed [24–29].In[30], Chen et al. proposed in-plane
rotation and scale invariant clustering using dictionaries. This
approach provides Radon-based rotation and scale invariant cluster -
ing as applied to content based image retrieval on Smithsonian
isolated leaf, Kimia shape and Brodatz texture datasets. Fei et al. [31]
described a CT image denoising based on sparse representation using
global dictionary. This approach impr ov ed lo w dose CT abdomen
image quality through a dictionary learning based denoising method
and accelerated the training time at the same time. Different classes
of images (produced by different departments such as dermatology
and pathology) were dealt with differently for applications such as
CBIR. An excellent review of the state-of-the-art of CBMIR and future
directions was presented in [32] . Several multi-resolution analysis
techniques via wa v elet, ridgelet, and curvelet-based textur e descrip-
tors were discussed for CBMIR [33]. The algorithm proposed therein
identified v arious tissues based on the discriminative texture features
with the aid of decision tree classification. This method too incorpo-
rated some tra ining data for realizing its objectives.
The present paper, nevertheless, has the objective of categoriz-
ing medical images that are not restricted to a specific context. In
applications of digital radiology such as computer aided diagnosis
or case based reasoning, the image category is of importance [3].It
may be emphasized here that our method
requires no training data for the classification (and retrieval) of
medical data, which is in contrast to existing methods
Fig. 1. Proposed feature extraction techniques: (a) image is partitioned into
concentric circular regions of equal area. (b) Image is divided into sub-images
and each sub-image is partitioned into concentric circular regions of equal area.
M. Srinivas et al. / Neurocomputing 168 (2015) 880–895 881



 我的内容管理
展开
我的内容管理
展开
 我的资源
快来上传第一个资源
我的资源
快来上传第一个资源
 我的收益 登录查看自己的收益
我的收益 登录查看自己的收益 我的积分
登录查看自己的积分
我的积分
登录查看自己的积分
 我的C币
登录后查看C币余额
我的C币
登录后查看C币余额
 我的收藏
我的收藏  我的下载
我的下载  下载帮助
下载帮助 
 前往需求广场,查看用户热搜
前往需求广场,查看用户热搜

 信息提交成功
信息提交成功