# PyPólyaGamma
[](https://travis-ci.org/slinderman/pypolyagamma)
This is a Cython port of Jesse Windle's code at
https://github.com/jwindle/BayesLogit. It provides a
Python interface for efficiently sampling Pólya-gamma
random variates. Install with:
pip install pypolyagamma
Please open issues if you have any trouble!
# Background
Pólya-gamma augmentation is a method of performing
fast and simple Bayesian inference in models with
Gaussian latent variables and count observations.
While such models are non-conjugate, if it has the
right form (specifically, if it is a Bernoulli, binomial,
negative binomial, or multinomial with a logistic link function),
we can introduce a set of Pólya-gamma
auxiliary variables that render it conditionally conjugate.
This facilitates fast Gibbs sampling algorithms on an
extended space of Gaussian latent variables
and Pólya-gamma auxiliary variables, where integrating out the
auxiliary variables leaves the original model intact.
Given the auxiliary variables, the latent Gaussian variables
have a Gaussian conditional distribution. Likewise, given
the Gaussian latent variables and the observed count data,
the auxiliary variables have a Pólya-gamma conditional distribution.
Thus, to implement the Gibbs sampling algorithm, we must be
able to efficiently sample Pólya-gamma random variates. This
library provides code to do exactly that.
The augmented density, the non-Gaussian marginal, and the Gaussian
conditionals are illustrated in the figure below. In this case, the posterior
is from a simple binomial model. Next, we'll show how to perform
Gibbs sampling for such a model.
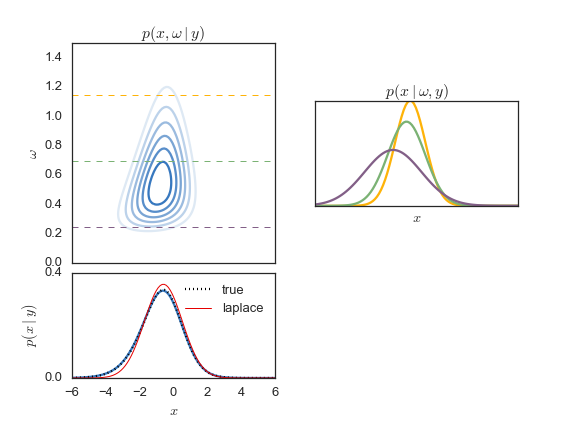
See below for more references and links.
# Demo
For convenience, we have created classes for simple
count regression models, like the Bernoulli, binomial, negative
binomial, and multinomial (with stick breaking) observation
models. For example, you can fit a Bernoulli regression as follows:
```python
from pypolyagamma import BernoulliRegression
D_out = 1 # Output dimension
D_in = 2 # Input dimension
reg = BernoulliRegression(D_out, D_in)
# Given X, an NxD_in array of real-valued inputs,
# and Y, and NxD_out array of binary observations. Fit
# the linear model y_n ~ Bern(sigma(A x_n + b)),
# where sigma is the logistic function. A is D_out x D_in,
# b is D_out x 1, and the entries in y_n are cond. indep.
samples = []
for _ in range(100):
reg.resample((X,Y))
samples.append((reg.A, reg.b))
```
Under the hood, this will instantiate Pólya-gamma auxiliary variables
and perform conditionally-conjugate Gibbs sampling. We can visualize
the inferred parameters in terms of the implied probability for each
point in the input space.
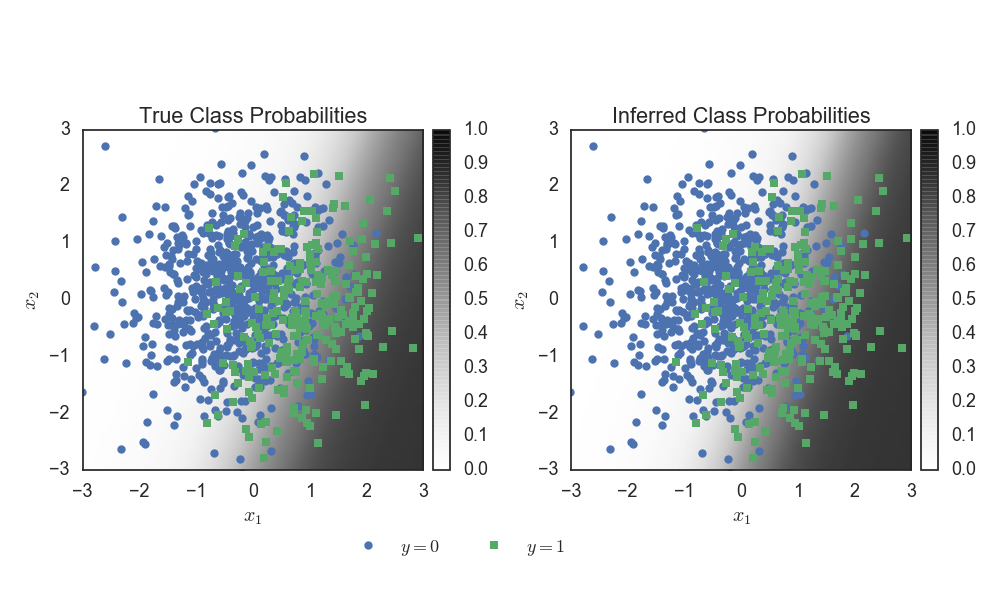
Here's how you can manually perform inference in a simple binomial model
with `N=10` counts and probability `p=logistic(x)`, with
a standard normal prior on `x`.
First, sample a count from the model:
```python
from pypolyagamma import logistic, PyPolyaGamma
# Consider a simple binomial model with unknown probability
# Model the probability as the logistic of a scalar Gaussian.
N = 10
mu = 0.0
sigmasq = 1.0
x_true = npr.normal(mu, np.sqrt(sigmasq))
p_true = logistic(x_true)
y = npr.binomial(N, p_true)
```
Now we can run a Gibbs sampler to estimate the posterior
distribution of `x` given `y`.
```python
# Gibbs sample the posterior distribution p(x | y)
# Introduce PG(N,0) auxiliary variables to render
# the model conjugate. First, initialize the PG
# sampler and the model parameters.
N_samples = 10000
pg = PyPolyaGamma(seed=0)
xs = np.zeros(N_samples)
omegas = np.ones(N_samples)
# Now run the Gibbs sampler
for i in range(1, N_samples):
# Sample omega given x, y from its PG conditional
omegas[i] = pg.pgdraw(N, xs[i-1])
# Sample x given omega, y from its Gaussian conditional
sigmasq_hat = 1./(1. / sigmasq + omegas[i])
mu_hat = sigmasq_hat * (mu / sigmasq + (y - N / 2.))
xs[i] = npr.normal(mu_hat, np.sqrt(sigmasq_hat))
```
For this simple example, we can compute the true posterior
and compare the samples to the target density.
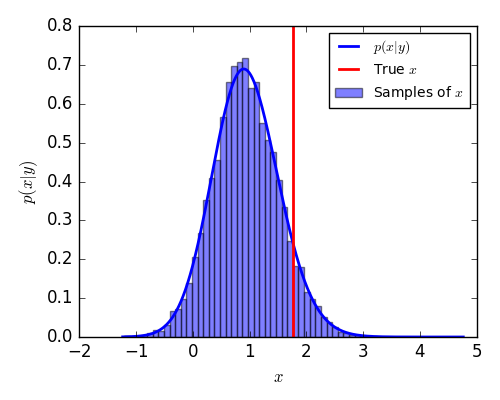
# Manual Installation
If you're a developer, you can also install from source:
git clone git@github.com:slinderman/pypolyagamma.git
cd pypolyagamma
pip install -e .
To check if it worked, run:
nosetests
If all the tests pass then you're good to go!
Under the hood, the installer will download
[GSL](https://www.gnu.org/software/gsl/),
untar it, and place it in `deps/gsl`. It will then configure GSL and
compile the Pólya-gamma code along with the required GSL source files.
This way, you don't need GSL to be installed and available on your
library path.
## Parallel sampling with OpenMP
By default, the simple installation above will not support
parallel sampling. If you are compiling with GNU `gcc` and `g++`,
you can enable OpenMP support with the flag:
USE_OPENMP=True pip install -e .
Mac users: you can install `gcc` and `g++` with Homebrew. Just
make sure that they are your default compilers, e.g. by setting
the environment variables `CC` and `CXX` to point to the GNU versions
of `gcc` and `g++`, respectively. With Homebrew, these versions
will be in `/usr/local/bin` by default.
To sample in parallel, call the `pgdrawvpar` method:
```python
n = 10 # Number of variates to sample
b = np.ones(n) # Vector of shape parameters
c = np.zeros(n) # Vector of tilting parameters
out = np.empty(n) # Outputs
# Construct a set of PolyaGamma objects for sampling
nthreads = 8
seeds = np.random.randint(2**16, size=nthreads)
ppgs = [pypolyagamma.PyPolyaGamma(seed) for seed in seeds]
# Sample in parallel
pypolyagamma.pgdrawvpar(ppgs, b, c, out)
```
If you haven't installed with OpenMP, this function will
revert to the serial sampler.
# References
- [Polson, Nicholas G., James G. Scott, and Jesse Windle. "Bayesian inference for logistic models using Pólya–Gamma latent variables." _Journal of the American statistical Association_ 108.504 (2013): 1339-1349.](http://www.tandfonline.com/doi/pdf/10.1080/01621459.2013.829001)
- [Windle, Jesse, Nicholas G. Polson, and James G. Scott. "Sampling Polya-Gamma random variates: alternate and approximate techniques." _arXiv preprint arXiv:1405.0506_ (2014).](http://arxiv.org/pdf/1405.0506)
- [Linderman, Scott, Matthew Johnson, and Ryan P. Adams. "Dependent Multinomial Models Made Easy: Stick-Breaking with the Polya-gamma Augmentation." _Advances in Neural Information Processing Systems (NIPS)_. 2015.](http://papers.nips.cc/paper/5660-dependent-multinomial-models-made-easy-stick-breaking-with-the-polya-gamma-augmentation.pdf) Check out our github repo, [pgmult](https://github.com/HIPS/pgmult)
- [Linderman, Scott W., Ryan P. Adams, and Jonathan W. Pillow. "Bayesian latent structure discovery from multi-neuron recordings." _Advances in Neural Information Processing Systems (NIPS)_ 2016.](https://arxiv.org/pdf/1610.08465) Check out our github repo, [pyglm](https://github.com/slinderman/pyglm))
- [Linderman, Scott W., Matthew Johnson, Andrew C. Miller, Ryan P. Adams, David M. Blei, and Liam Paninski. "Bayesian learning and inference in recurrent switching linear dynamical systems." _Artificial Intelligence and Statistics (AISTATS)_ 2017.](https://arxiv.org/pdf/1610.08466.pdf)
 pypolyagamma-1.1.5.tar.gz (37个子文件)
pypolyagamma-1.1.5.tar.gz (37个子文件)  pypolyagamma-1.1.5
pypolyagamma-1.1.5  MANIFEST.in 262B
MANIFEST.in 262B PKG-INFO 578B
PKG-INFO 578B pypolyagamma
pypolyagamma  parallel.pyx 1KB
parallel.pyx 1KB utils.py 5KB
utils.py 5KB cpp
cpp  PolyaGammaAlt.h 3KB
PolyaGammaAlt.h 3KB PolyaGammaAlt.cpp 7KB
PolyaGammaAlt.cpp 7KB PolyaGammaSP.cpp 6KB
PolyaGammaSP.cpp 6KB include
include  GRNG.hpp 3KB
GRNG.hpp 3KB RNG.cpp 11KB
RNG.cpp 11KB RNG.hpp 6KB
RNG.hpp 6KB GRNG.cpp 4KB
GRNG.cpp 4KB SRNG.hpp 3KB
SRNG.hpp 3KB PolyaGamma.h 2KB
PolyaGamma.h 2KB InvertY.cpp 2KB
InvertY.cpp 2KB PolyaGammaSmallB.h 731B
PolyaGammaSmallB.h 731B PolyaGammaSmallB.cpp 2KB
PolyaGammaSmallB.cpp 2KB PolyaGammaOMP.h 2KB
PolyaGammaOMP.h 2KB PolyaGamma.cpp 6KB
PolyaGamma.cpp 6KB PolyaGammaSP.h 1KB
PolyaGammaSP.h 1KB PolyaGammaPar.h 8KB
PolyaGammaPar.h 8KB PolyaGammaHybrid.h 2KB
PolyaGammaHybrid.h 2KB InvertY.hpp 2KB
InvertY.hpp 2KB distributions.py 23KB
distributions.py 23KB parallel.cpp 731KB
parallel.cpp 731KB __init__.py 865B
__init__.py 865B pypolyagamma.pyx 987B
pypolyagamma.pyx 987B pypolyagamma.cpp 693KB
pypolyagamma.cpp 693KB deps
deps  README.md 226B
README.md 226B test
test  test_basics.py 3KB
test_basics.py 3KB setup.cfg 79B
setup.cfg 79B setup.py 6KB
setup.py 6KB pypolyagamma.egg-info
pypolyagamma.egg-info  PKG-INFO 578B
PKG-INFO 578B requires.txt 23B
requires.txt 23B SOURCES.txt 1KB
SOURCES.txt 1KB top_level.txt 13B
top_level.txt 13B dependency_links.txt 1B
dependency_links.txt 1B README.md 8KB
README.md 8KB
 我的内容管理
展开
我的内容管理
展开
 我的资源
快来上传第一个资源
我的资源
快来上传第一个资源
 我的收益 登录查看自己的收益
我的收益 登录查看自己的收益 我的积分
登录查看自己的积分
我的积分
登录查看自己的积分
 我的C币
登录后查看C币余额
我的C币
登录后查看C币余额
 我的收藏
我的收藏  我的下载
我的下载  下载帮助
下载帮助 
 前往需求广场,查看用户热搜
前往需求广场,查看用户热搜

 信息提交成功
信息提交成功